|
|
 |
Back to Annual Meeting Program
Expression Analysis of Macrodactyly Identifies Pleiotrophin Upregulation
Frank Lau, MD1, Fang Xia2, Adam Kaplan2, Felecia Cerrato1, Arin K. Greene, MD1, Amir Taghinia, MD1, Chad A. Cowan, PhD2, Brian I. Labow, MD1.
1Children's Hospital Boston, Boston, MA, USA, 2Massachusetts General Hospital, Boston, MA, USA.
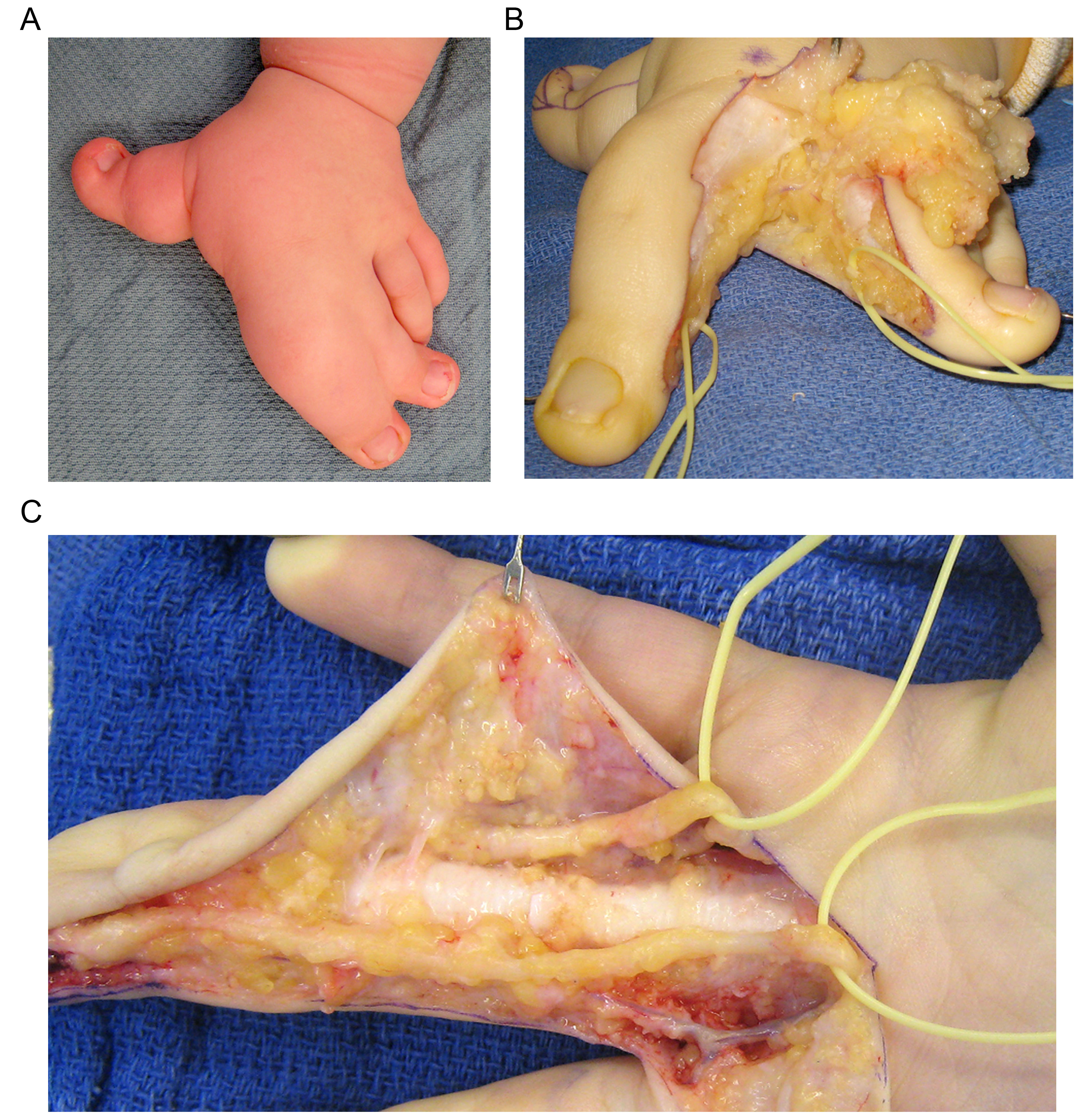
BACKGROUND:Macrodactyly is a rare family of congenital disorders characterized by the diffuse enlargement of 1 or more digits. Multiple tissue types within the affected digits are involved, but skeletal patterning and gross morphological features are preserved. Not all tissues are equally involved and there is marked heterogeneity with respect to clinical phenotype. The molecular mechanisms responsible for these growth disturbances offer unique insight into normal limb growth and development, in general. To date, no genes or loci have been implicated in the development of macrodactyly.
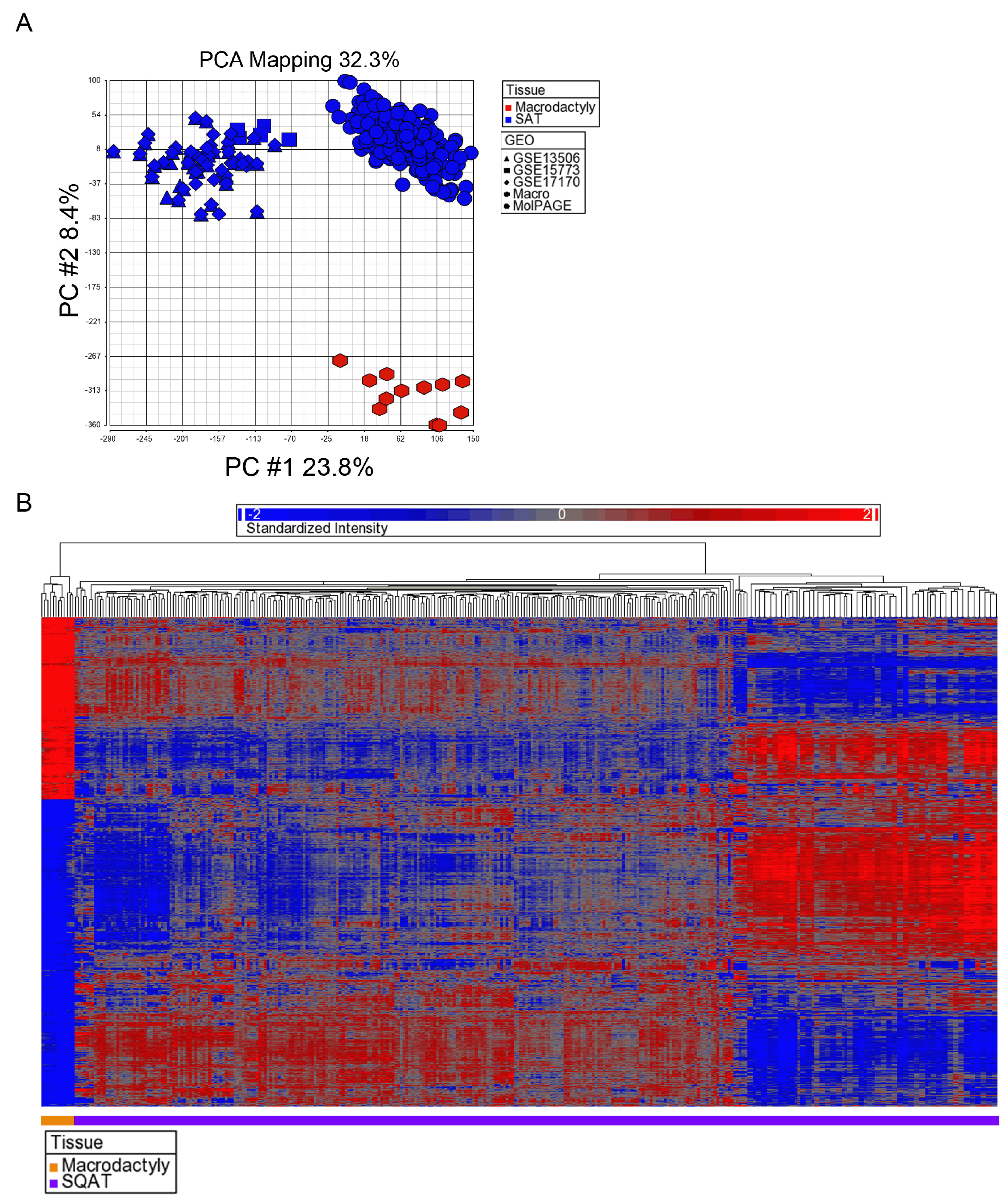
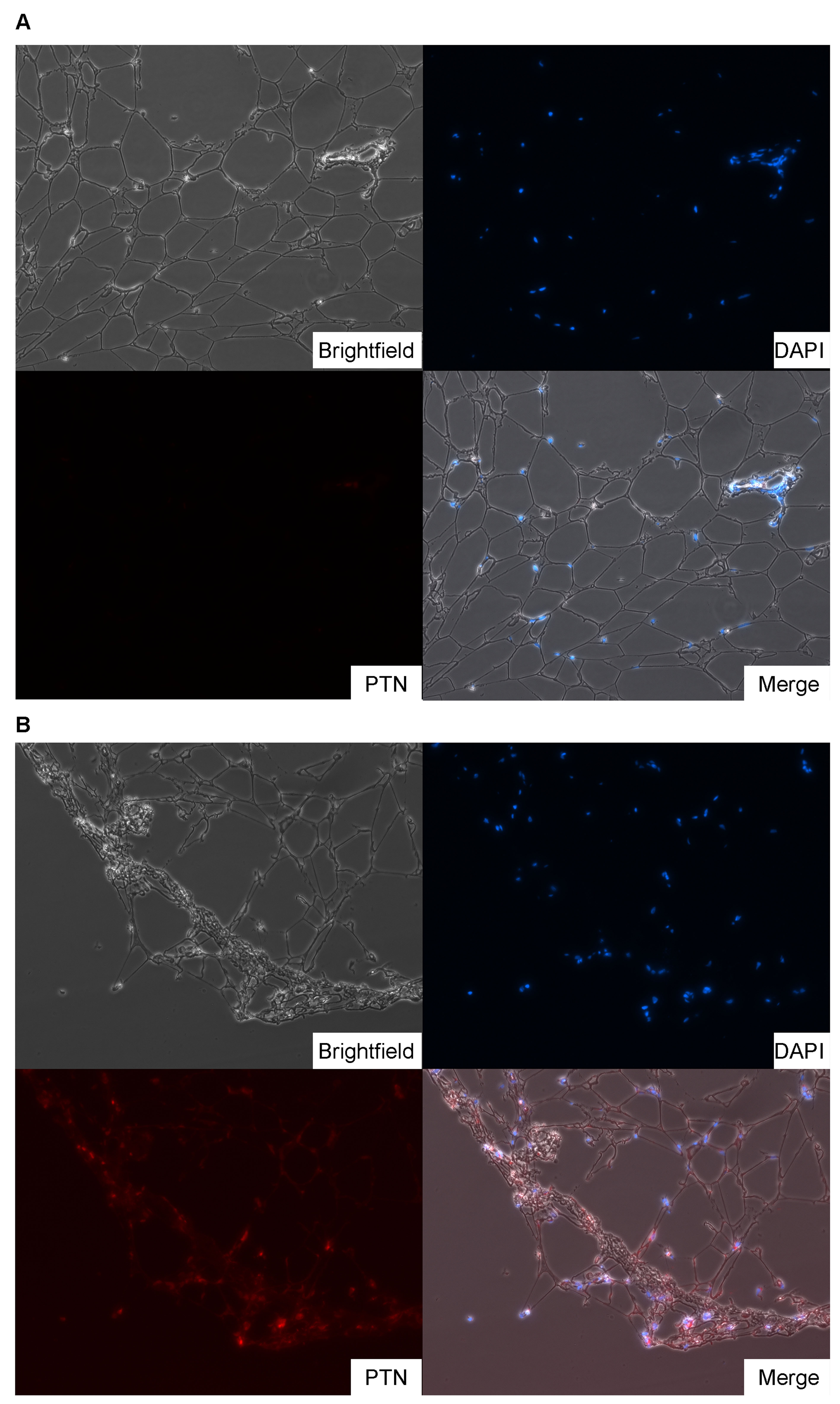
METHODS: In this study, we performed transcriptional profiling of macrodactyly tissue. RNA was isolated from 4 macrodactyly adipose tissue samples and hybridized to Affymetrix U133A Plus 2.0 microarrays. Microarray data were analyzed with principal component analysis, hierarchical clustering, and gene enrichment analysis. Significant results were confirmed using quantitative real time polymerase chain reaction and immunohistochemistry.
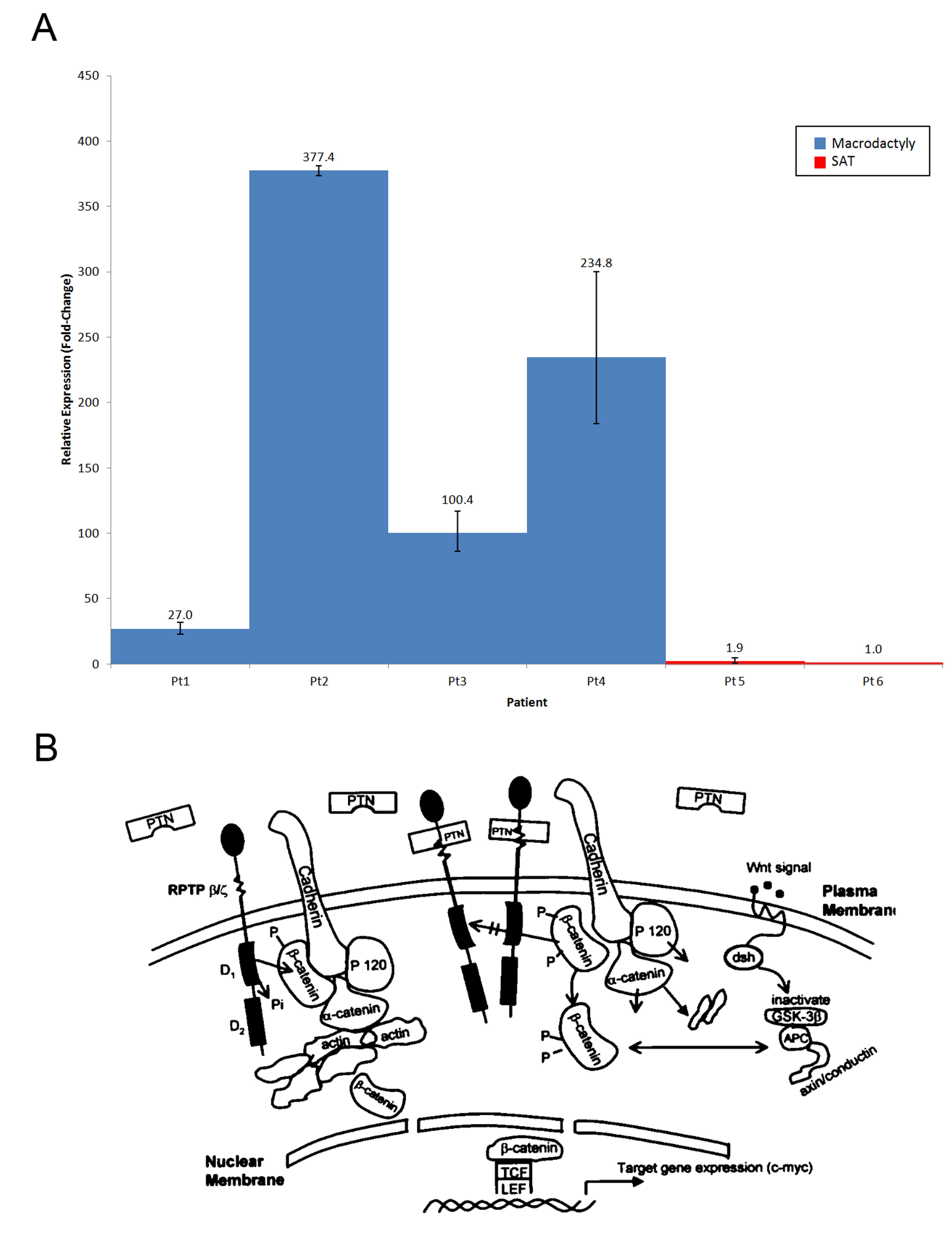
RESULTS: 7,295 genes are differentially expressed in macrodactyly adipose tissue compared to subcutaneous adipose tissue. In particular, pleiotrophin (PTN) was 127.6-fold (p-value = 0.049) overexpressed across all our macrodactyly samples. At the biological pathway level, gene enrichment analysis found that subcutaneous adipose tissue (SAT) was enriched for classic adipose tissue gene ontology categories such as “regulation of catabolic processes” and “response to insulin stimulus”. In contrast, macrodactyly tissues demonstrated enrichment of vastly different gene ontology categories such as “extracellular space” and “pattern binding”.
CONCLUSIONS:Pleiotrophin is a promising candidate gene for the pathogenesis of macrodactyly because it promotes growth of nearly all the tissues affected by macrodactyly, including nerve, skin, bone, and cartilage. PTN thus represents a promising target for further investigation into the etiology of overgrowth phenotypes.
Top 10 greatest fold-change probesets in macrodactyly| Probeset ID | Gene | Description | p-value | Fold-Change (vs. subcutaneous adipose tissue) | | 209465_x_at | PTN | pleiotrophin | 0 | 34.44 | | 223475_at | CRISPLD1 | cysteine-rich secretory protein LCCL domain containing 1 | 1.85E-42 | 21.42 | | 211737_x_at | PTN | pleiotrophin | 0 | 19.86 | | 220504_at | KERA | keratocan | 0 | 18.51 | | 203913_s_at | HPGD | hydroxyprostaglandin dehydrogenase 15-(NAD) | 1.40E-45 | 15.71 | | 216834_at | RGS1 | regulator of G-protein signaling 1 | 2.72E-25 | 14.19 | | 205430_at | BMP5 | bone morphogenetic protein 5 | 0 | 13.97 | | 209466_x_at | PTN | pleiotrophin | 0 | 12.97 | | 209189_at | FOS | FBJ murine osteosarcoma viral oncogene homolog | 3.03E-15 | 12.82 |
Top 10 greatest fold-change probesets in subcutaneous adipose tissue| Probeset ID | Gene | Description | p-value | Fold-Change (vs. Macrodactyly) | | 214456_x_at | SAA1 /// SAA2 | serum amyloid A1 /// serum amyloid A2 | 4.20E-45 | 91.68 | | 208607_s_at | SAA1 /// SAA2 | serum amyloid A1 /// serum amyloid A2 | 4.84E-26 | 72.59 | | 204424_s_at | LMO3 | LIM domain only 3 (rhombotin-like 2) | 0 | 17.56 | | 214146_s_at | PPBP | pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | 1.72E-21 | 16.27 | | 228434_at | BTNL9 | butyrophilin-like 9 | 0 | 14.31 | | 211699_x_at | HBA1 /// HBA2 | hemoglobin, alpha 1 /// hemoglobin, alpha 2 | 0 | 14.28 | | 209458_x_at | HBA1 /// HBA2 | hemoglobin, alpha 1 /// hemoglobin, alpha 2 | 0 | 13.49 | | 204018_x_at | HBA1 /// HBA2 | hemoglobin, alpha 1 /// hemoglobin, alpha 2 | 0 | 12.66 | | 204105_s_at | NRCAM | neuronal cell adhesion molecule | 2.39E-27 | 12.49 |
Enriched gene ontology categories in macrodactyly| # of genes | P-adjusted | GO ID | GO Category | | 6 | 0.001 | GO:0071363 | cellular response to growth factor stimulus | | 6 | 0.039 | GO:0070848 | response to growth factor stimulus | | 10 | 0.048 | GO:0030199 | collagen fibril organization | | 18 | 0.011 | GO:0001501 | skeletal system development | | 19 | 0 | GO:0005578 | proteinaceous extracellular matrix | | 22 | 0 | GO:0031012 | extracellular matrix | | 30 | 0 | GO:0005539 | glycosaminoglycan binding | | 34 | 0 | GO:0001871 | pattern binding | | 34 | 0 | GO:0030247 | polysaccharide binding |
Enriched gene ontology categories in subcutaneous adipose tissue| # of genes | P-adjusted | GO ID | GO Category | | 3 | 0.002 | GO:0015671 | oxygen transport | | 12 | 0.013 | GO:0003995 | acyl-CoA dehydrogenase activity | | 17 | 0 | GO:0046320 | regulation of fatty acid oxidation | | 24 | 0.023 | GO:0032869 | cellular response to insulin stimulus | | 36 | 0.004 | GO:0032868 | response to insulin stimulus | | 43 | 0 | GO:0001525 | angiogenesis | | 55 | 0.013 | GO:0051056 | regulation of small GTPase mediated signal transduction | | 60 | 0.049 | GO:0006732 | coenzyme metabolic process | | 87 | 0.004 | GO:0009894 | regulation of catabolic process |
Back to Annual Meeting Program
|










