Back to 2015 Annual Meeting
Identification of new gene mutations in Lymphangioma patients
Mark Youngblood, Brandon Sumpio, Andre Alcon, Soonwook Hong, Murat Gunel, Deepak Narayan.
Yale School of Medicine, New Haven, CT, USA.
Introduction:
Lymphatic malformations are rare, sporadic congenital lesions found in lymphatic-rich tissues. During normal embryonic development, lymphatic endothelial cells bud from anterior cardinal veins. The budding is driven primarily by Prox1, which stimulates expression of other lymphatic developmental genes such as LYVE1 and VEGFC; constant Prox-1 expression helps lymphatic endothelial cells maintain tier phenotype.
While lymphatic remodeling is done constantly in utero, 1 in 250 infants show nuchal translucency an indicator of aberrant lymphatic development,. However, 1 in 5000 newborns have lymphatic malformations. Thus the remodeling must occur in utero. This paper examines the genes associated with the lymphangiomas and classifies subsets of genes that may be important in downstream regulation.
Methods:
Eight lymphangioma and matching blood samples underwent whole-exome sequencing (WES) to identify somatic driver mutations. Data was pre-processed to remove low-quality reads and NGS adaptors, aligned using BWA, and further processed with MarkDuplicates (Picard) and BaseQualityScoreRecalibration (GATK). Variant calling was performed with HaplotypeCaller (GATK), and annotated using Annovar. The results of this discovery cohort were validated in an independent group of 30 samples using molecular-inversion probe sequencing. This provided high-coverage, targeted reads of genomic areas of interest identified in WES. To investigate the involvement of aberrant gene expression, mRNA sequencing was performed on 6 tumor samples. After pre-processing, data was aligned using TopHat2 and differential expression analysis was performed with Cuffdiff.
Results:
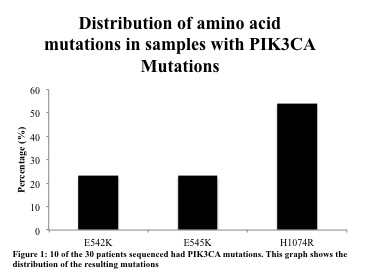
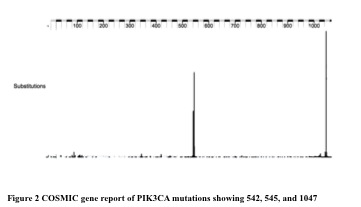
WES identified recurrent alterations to the COSMIC reported gene PIK3CA in three of eight samples. PI3k is an intracellular signaling pathway important for regulating cell cycle and cell proliferation. High-coverage targeted re-sequencing confirmed the variants in these three samples, and identified ten additional samples with PIK3CA mutations out of thirty examined (Fig 1) 23% (3 out of the 13 PIK3CA mutated samples) had a mutation amino acid mutation changing Glutamic acid (E) for lysine at 542 and another 23% at 545. The majority was of the mutations 54% occurred at 1047 resulting in a Histidine switch for Arginine. As shown in figure 2, these mutations occur at similar COSMIC hotspots in the gene, and cause the same E542K, E545K, and H1047R alterations in the resulting protein.
Back to 2015 Annual Meeting
|




