T-Cell Clonality in Lymphedema: Linking Pathogenic T-cell Clones with Disease
Adana Campbell1, Jungeun Baik1. Babak Mehrara1
Memorial Sloan Kettering, New York, NY
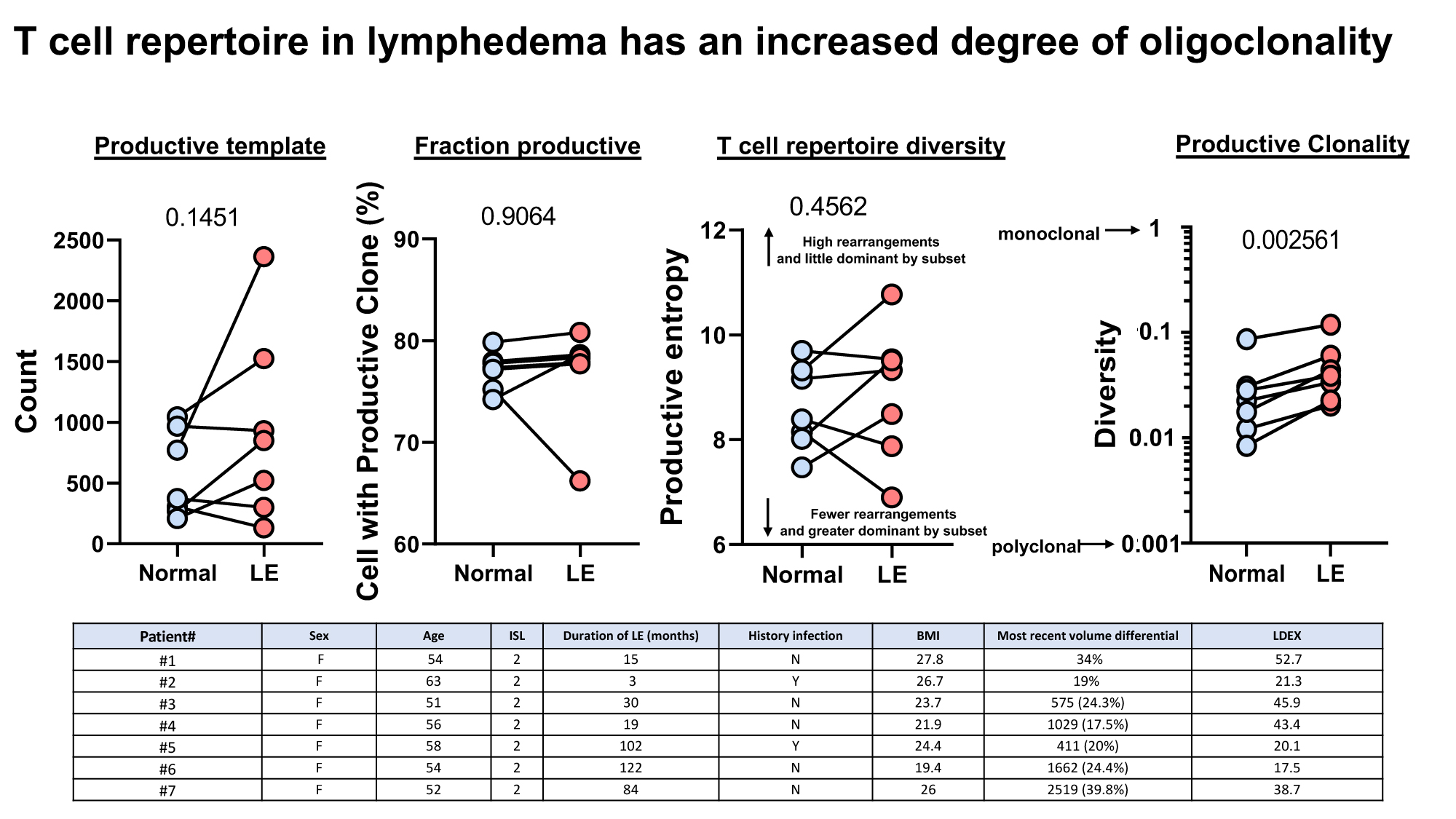
Lymphedema (LE) is a progressive condition that is estimated to affect as many as 1 in 1000 Americans. Activation of T-helper (Th) inflammatory responses plays a key role in lymphedema development and has been correlated with disease severity in LE mouse models. This activation requires interaction with antigen-presenting cells (APCs), suggesting that the T-cell responses observed occur because of antigens that are present in tissues with lymphatic fluid stasis. Clonal analysis of these T cell responses is significant due to its potential to uncover a common clonal architecture between patients with the disease. Identification of antigen-responsive T cell clones that are selectively expanded will change the way lymphedema is studied and has important implications for developing therapies aimed at a common antigen driving the disease process. Genomic DNA (gDNA) was isolated from paired, lymphedema and normal, upper limb biopsies of six patients and subject to high throughput sequencing (HTS). Deep sequencing of the TCR repertoire was conducted by Adaptive Biotechnologies using a two-step biased controlled multiplex PCR to amplify genes of abundant T cell populations, and clones were quantified and characterized by CDR3 sequence length and TCRB Variable (V) gene usage. Predictive analysis of T cell antigen specificity was performed using the Basic Local Alignment Search Tool (BLAST) of the TCR epitope sequences and the activation of antigen-responsive clones was assessed by Flow cytometry. The T cell responses in lymphedema are oligoclonal. TCR sequencing of the top five abundant clones in lymphedematous and normal tissue biopsies revealed six conserved amino acid sequences at the CDR3 region unique to the lymphedema group, however, no overlap of T cell repertoires was seen between patients with the disease. BLAST analysis of the corresponding TCRB epitope sequence revealed a list of probable antigens corresponding to each unique clonotype. Of the epitope sequences, unique to LE, staphylococcal enterotoxin represented the highest affinity bacterial antigen, pro-insulin corresponded to the representative self-antigen, and gluten peptide corresponded to a plant antigen. Our results indicate that the T cell response in lymphedema is oligoclonal, similar to what is seen in other inflammatory skin diseases such as atopic dermatitis. Clonality infers an antigen-driven T-cell response. Here we link a group of putative antigens driving this clonal response. Identification of these T cell populations that are selectively expanded in LE may provide a targeted approach for identifying and eliminating pathogenic T cell clones as a means of treating the disease.
Back to 2022 Abstracts